New insights into the mechanisms of human coronaviruses revealed
Scientists have discovered new details about how coronaviruses make copies of themselves inside human cells. They studied how the virus splits long protein chains into smaller parts, which then come together to form the virus’s replication machine. Understanding this process better can help develop new treatments to stop the virus from spreading.
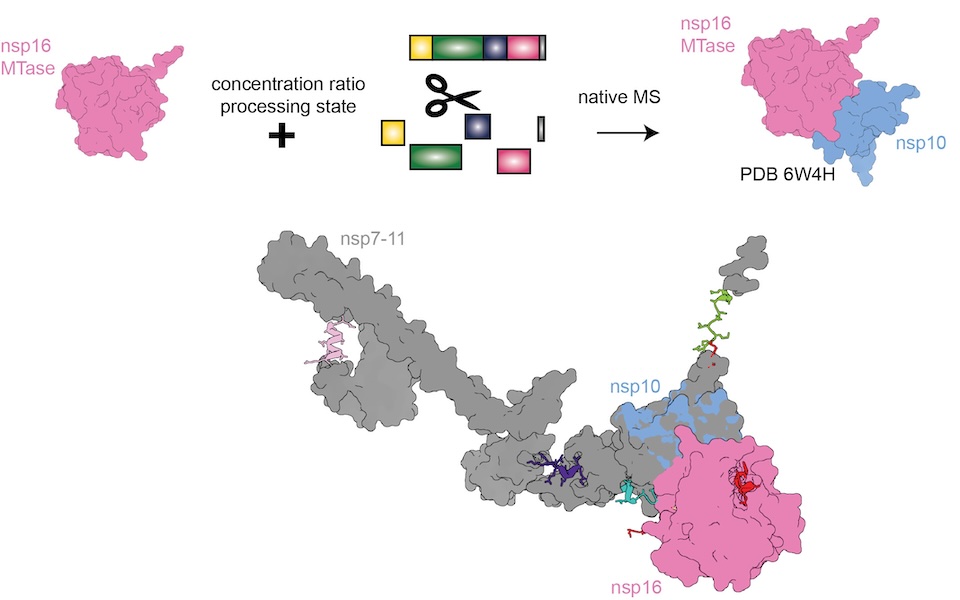
To date, there are seven different coronaviruses (CoV) that are known to infect humans. These viruses range from HCoV-229 which causes mild colds to more virulent pathogens such as SARS-CoV-2, which has caused over 7.1 million deaths since 2019. All human coronaviruses contain a single stranded RNA genome and share many similarities in viral replication. The group of Charlotte Uetrecht (University Lübeck and DESY) has discovered new insights into the order of polyprotein processing and its influence on the formation of the replication/transcription complex (RTC). Their findings are now published in Nature Communications.
To propagate infection, viruses need to replicate inside their human host. The coronavirus uses its spike proteins to attach to a human cell and bring about the fusion of the viral and cellular membranes. Once a coronavirus’ viral genome has entered its human host, the cellular ribosome meets viral RNA and begins.
A string of pearls
“One can think of polyproteins as a string of pearls where each individual pearl is a nonstructural protein (nsp)” explains the paper’s first author Kira Schamoni-Kast “together the pearls are flexible and they have a favorite conformation, or way of folding, in the space they are occupying. However, this is an intermediate stage and it is only when the individual proteins, are cut from the string that they are able to interact with other parts of the host cell and continue the viral replication process.”
Schamoni-Kast and her colleagues were interested in understanding whether the order in which each nsp is cleaved from the string by the viral main protease Mpro influences the assembly of the RTC. The team focused on a determining the processing mechanisms of a particular set of nsps, namely the nsp7-11 region, which is known to be crucial for viral growth.
Given the flexible nature of polyproteins, there was little high resolution structural data available to the researchers. The team turned to native mass spectrometry (MS), a highly sensitive method that acts a bit like a molecular scale and maintains native-like protein folding. This allowed for the simultaneous monitoring of the polyprotein cleavage products. “Native MS enabled us to study this dynamic process,” notes corresponding author Charlotte Uetrecht. “Native MS not only preserved protein-protein interactions but it also allowed us to extract cleavage site kinetics and detect of protein complexes formed by the processing products.”
The importance of processing order
By determining the rate constants k, which indicates the speed of a chemical reaction, from nsp cleavage sites, the researchers revealed the processing of nsp7-11 from four different human pathogenic CoV species and determined distinct mechanisms for each cleavage site based on the local structural environments. “Our comparative analysis revealed both conserved and unique features in nsp7-11 processing reactions of the four human CoV species. We also demonstrated that while complete processing of nsp7-11 is not essential, it greatly enhances complex assembly,” explains Uetrecht.
In fact, the cleavage rate is consistently delayed at cleavage site 7/8 across all species of CoV which hints at a possible regulatory mechanism for the assembly of coronavirus polymerase complexes. “I like to use the analogy of a family getting ready for a summer vacation,” explains Schamoni-Kast “nsp12, as polymerase, is the busy parent running around and packing the car. Nsp7 and nsp8 are the children who are put into the car at the last possible moment after everything else has been packed. Once everyone is in the car, the family can take off for their vacation destination.” This sequence demonstrates how the spatiotemporal coordination of nsps is likely to help orchestrate the sequential formation of various functional assemblies. It would, for example, ensure that RNA capping and proof-reading are in place first which would subsequently allow the virus to fine-tune RNA synthesis and modification.
“Overall, an enhanced understanding of polyprotein processing will aid in the identification of rate-limiting steps and critical interaction sites that could become the targets of future therapeutic interventions,” notes Uetrecht.
Original publication
Schamoni-Kast K, Krichel B, Damjanović T, Said F-A, Kierspel T, Toker S, Uetrecht C (2025) The kinetics of nsp7-11 polyprotein processing and impact on complexation with nsp16 among human coronaviruses. Nat Commun 16, 8244
