Example (GAMESSUS): Sampling initial conditions for a water molecule
This example shows you how to generate an ensemble of initial conditions (atom positions and velocities) for a single water molecule. Input files can be found in the directory:
Examples/xsample/
Preparation
You need to first generate a Hessian file containing the information about the derivatives of the potential energy surface.
In the current example, this has been done using a GAMESSUS calculation.
The input file that was used for GAMESSUS is provided as h2o_6-31g_hess.inp.
The GAMESSUS calculation results in the file h2o_6-31g_hess.dat.
Using:
an_getHessian -H h2o_6-31g_hess.dat > hessian
the hessian information is extracted from the GAMESSUS output file (in fact the “.dat” file) into the file hessian
Running
To run the sampling, execute:
xsample -i input_h2o_sampling -d h2o_sampling -H hessian
Input file
The file input_h2o_sampling is the input file used for the sampling.:
$SYSTEM
qchemistry = gamess
xunit = an
rseed = 1
$END
$sampling
type = wignerHarmonic
# type = zero_point_energy
nsample = 10
modes = nonlinear
$end
$cartPOS
O 0.0000000000 -0.0000000000 0.0349530522
H 0.0000000000 0.7852216674 0.5690592739
H 0.0000000000 -0.7852216674 0.5690592739
$end
$cartVEL
O 0.0 0.0 0.0
H 0.0 0.0 0.0
H 0.0 0.0 0.0
$end
The following input parameters have been used:
$SYSTEMdescribes general parameters for the samplingThe option
xunit = anspecifies the coordinate units. “an” refers to Angstrom.The option
qchemistry = gamessspecifies GAMESSUS as the quantum chemistry program
The section
$samplingdefines the type of samplingtype can be
zero_point_energyorwignerHarmonicnsamplegives the number of samplesmodesspecifies the virbrational normal modes that are used for samplingmodes = nonlinearskips the lowest 3N-6 modesmodes = linearskips the lowest 3N-5
$cartPOSspecifies the geometry from which the sampling is generated. For Wigner sampling based on harmonic approximation the position should be the energy minimum. In the current example energy minimization has been performed for the water molecule and the virbrational constants have been calculated using GAMESSUS, see the fileh2o_6-31g_hess.inp$cartVELspecifies the velocities at which the sampling is preformed. Usually they are zero.The file
hessiancontains the Hessian matrix data that has been extracted from the GAMESSUS calculation. See the fileh2o_6-31g_hess.inp
Output results
xsample creates a directory ensmbl_h2o_sampling with subdirectories named trj_n, where n runs from 0 to the number of samples. Each trajectory contains files storing position and velocity information for each atom that can be used to initiate trajectories with xpyder.
You may inspect the initial geometries by converting them into the common xyz file format. To do so, you may execute:
for d in trj_*; do cd $d; cdtk2xyz ; cd ..; done
This creates for in each trj_n subdirectory a file geom.xyz.
You can inspect the initial geometries via vmd:
vmd -m trh_*/geom.xyz
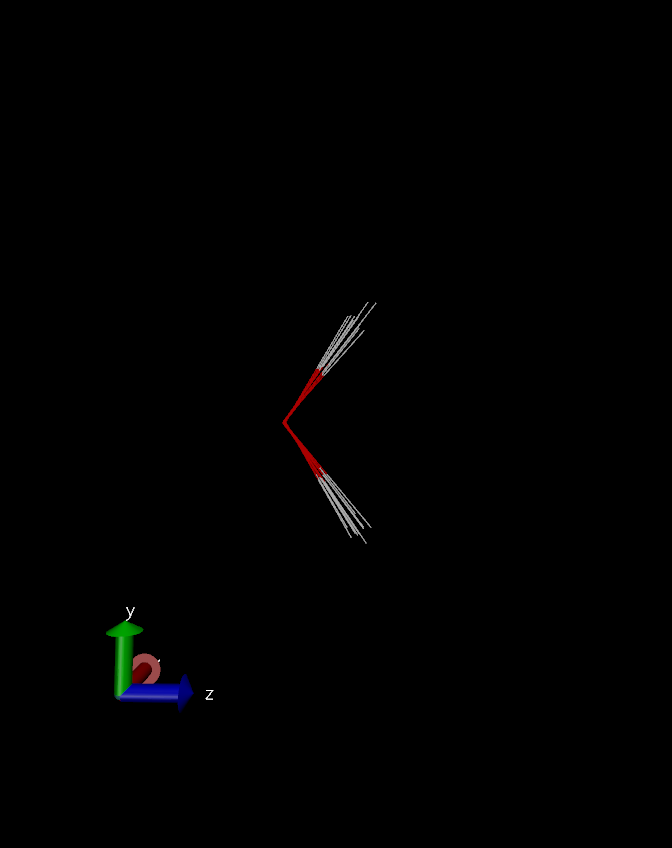
Overlayed water geometries depicting zero-point fluctuations.